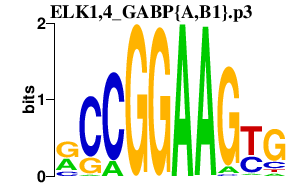
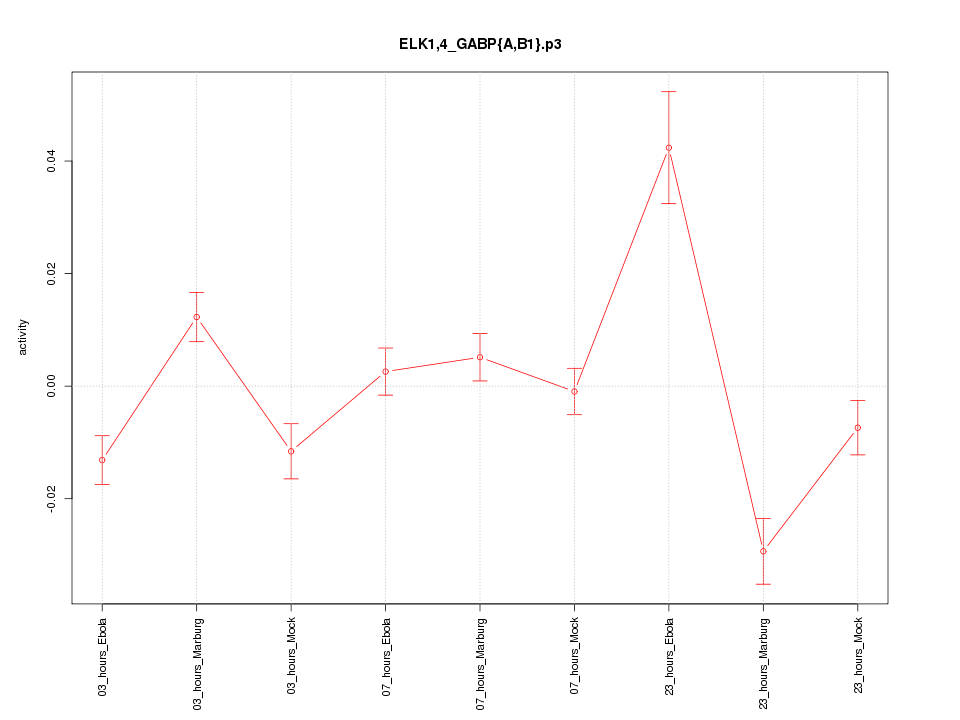
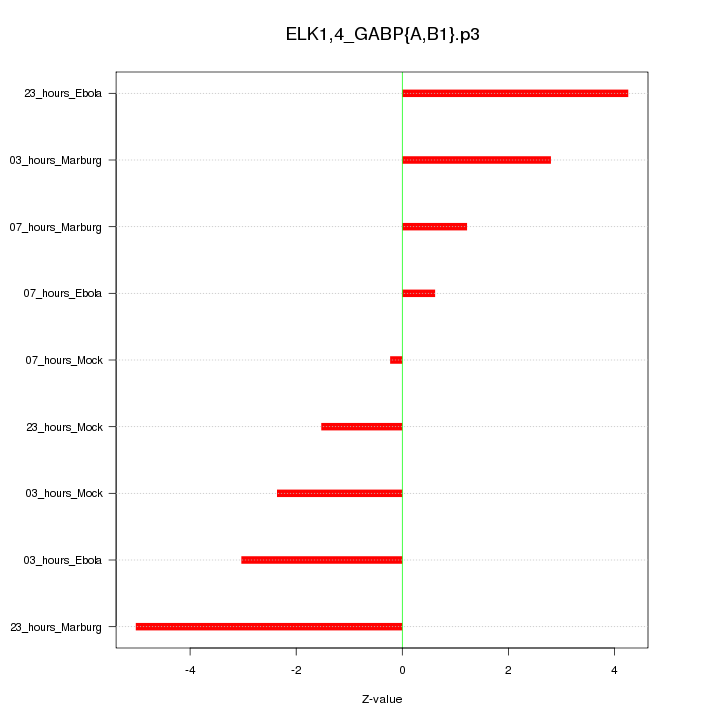
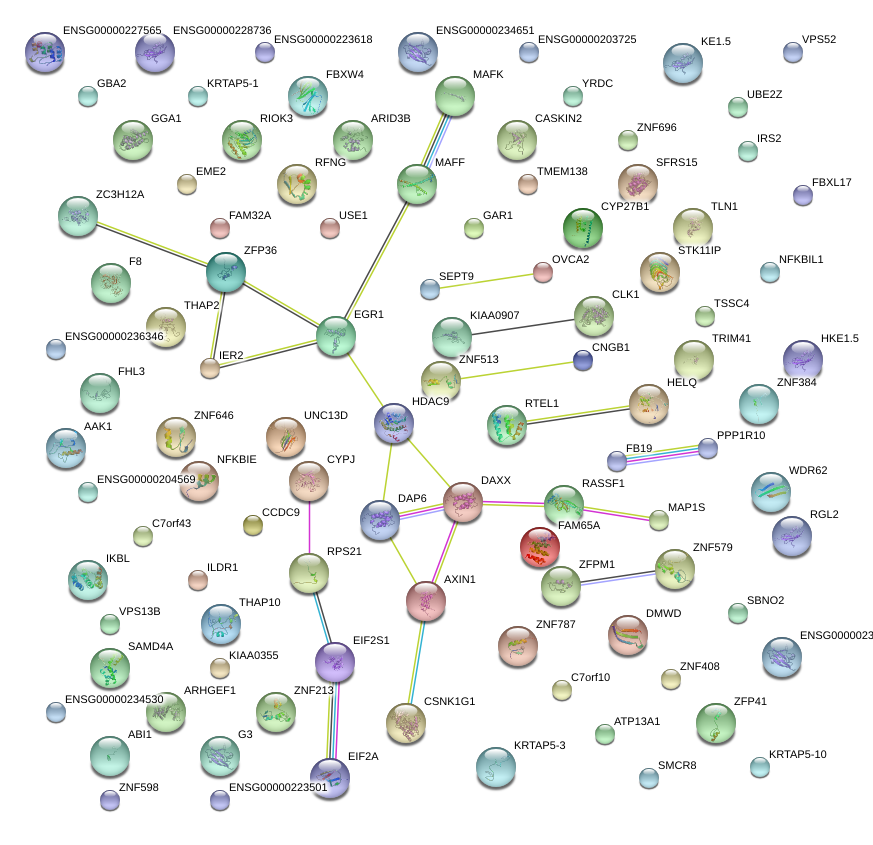
|
chr5_+_137801178
|
3.889
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr5_+_137801166
|
3.882
|
|
EGR1
|
early growth response 1
|
|
chr11_+_2421983
|
3.109
|
|
TSSC4
|
tumor suppressing subtransferable candidate 4
|
|
chr11_+_46722564
|
2.976
|
NM_001184751
|
ZNF408
|
zinc finger protein 408
|
|
chr11_+_46722316
|
2.797
|
NM_024741
|
ZNF408
|
zinc finger protein 408
|
|
chr6_+_31515352
|
2.698
|
NM_001144961
NM_005007
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1
|
|
chr12_-_58159386
|
2.665
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr3_-_50378238
|
2.504
|
NM_001206957
NM_007182
NM_170714
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr8_+_144349564
|
2.491
|
NM_138465
|
GLI4
|
GLI family zinc finger 4
|
|
chr16_+_31085722
|
2.465
|
NM_014699
|
ZNF646
|
zinc finger protein 646
|
|
chr19_+_47759802
|
2.353
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr2_-_27603249
|
2.253
|
NM_001201459
|
ZNF513
|
zinc finger protein 513
|
|
chr7_-_99756329
|
2.234
|
|
C7orf43
|
chromosome 7 open reading frame 43
|
|
chr16_-_402617
|
2.227
|
NM_003502
NM_181050
|
AXIN1
|
axin 1
|
|
chr16_+_31085751
|
2.223
|
|
ZNF646
|
zinc finger protein 646
|
|
chr15_+_74833517
|
2.171
|
NM_006465
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr7_-_99756275
|
2.170
|
NM_018275
|
C7orf43
|
chromosome 7 open reading frame 43
|
|
chr19_+_47759730
|
2.153
|
NM_015603
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr5_-_107717072
|
2.141
|
|
FBXL17
|
F-box and leucine-rich repeat protein 17
|
|
chr22_+_38599026
|
2.129
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr19_-_56092184
|
2.092
|
NM_152600
|
ZNF579
|
zinc finger protein 579
|
|
chr16_-_2059759
|
2.052
|
NM_178167
|
ZNF598
|
zinc finger protein 598
|
|
chr15_-_64648359
|
2.042
|
NM_022048
|
CSNK1G1
|
casein kinase 1, gamma 1
|
|
chr19_+_42387249
|
2.033
|
NM_004706
NM_198977
|
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1
|
|
chr15_-_71184611
|
2.032
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr5_+_180650274
|
2.010
|
|
TRIM41
|
tripartite motif containing 41
|
|
chr1_-_38273823
|
2.009
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr19_+_16296211
|
2.004
|
NM_014077
|
FAM32A
|
family with sequence similarity 32, member A
|
|
chr6_-_30585016
|
1.993
|
NM_002714
|
PPP1R10
|
protein phosphatase 1, regulatory subunit 10
|
|
chr11_-_1606480
|
1.982
|
NM_001005922
|
KRTAP5-1
|
keratin associated protein 5-1
|
|
chr14_+_55034329
|
1.972
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr5_+_180650233
|
1.969
|
NM_033549
NM_201627
|
TRIM41
|
tripartite motif containing 41
|
|
chr6_-_30585024
|
1.968
|
|
PPP1R10
|
protein phosphatase 1, regulatory subunit 10
|
|
chr2_-_27603584
|
1.962
|
NM_144631
|
ZNF513
|
zinc finger protein 513
|
|
chr19_+_17326196
|
1.957
|
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae)
|
|
chr6_-_33267128
|
1.943
|
NM_001243738
NM_004761
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr19_-_19774472
|
1.941
|
|
ATP13A1
|
ATPase type 13A1
|
|
chr16_-_2059795
|
1.940
|
|
ZNF598
|
zinc finger protein 598
|
|
chr19_-_19774501
|
1.907
|
NM_020410
|
ATP13A1
|
ATPase type 13A1
|
|
chr19_+_13261272
|
1.903
|
NM_004907
|
IER2
|
immediate early response 2
|
|
chr1_+_37940074
|
1.885
|
NM_025079
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr17_+_78388944
|
1.879
|
NM_001164637
NM_001164638
NM_173627
|
ENDOV
|
endonuclease V
|
|
chr16_+_3185112
|
1.873
|
NM_001134655
|
ZNF213
|
zinc finger protein 213
|
|
chr19_-_56632644
|
1.839
|
NM_001002836
|
ZNF787
|
zinc finger protein 787
|
|
chr1_+_156698270
|
1.823
|
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1
|
|
chr2_+_220462560
|
1.818
|
NM_052902
|
STK11IP
|
serine/threonine kinase 11 interacting protein
|
|
chr16_+_1823184
|
1.809
|
NM_001010865
|
EME2
|
essential meiotic endonuclease 1 homolog 2 (S. pombe)
|
|
chr4_+_110736636
|
1.794
|
NM_018983
NM_032993
|
GAR1
|
GAR1 ribonucleoprotein homolog (yeast)
|
|
chr20_+_60962120
|
1.786
|
NM_001024
|
RPS21
|
ribosomal protein S21
|
|
chr17_-_73505914
|
1.771
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr8_+_100025501
|
1.770
|
|
VPS13B
|
vacuolar protein sorting 13 homolog B (yeast)
|
|
chr8_+_100025493
|
1.741
|
NM_015243
NM_017890
NM_152564
NM_181661
|
VPS13B
|
vacuolar protein sorting 13 homolog B (yeast)
|
|
chr16_-_402398
|
1.736
|
|
AXIN1
|
axin 1
|
|
chr19_-_1174263
|
1.712
|
NM_014963
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr9_-_35749144
|
1.706
|
|
GBA2
|
glucosidase, beta (bile acid) 2
|
|
chr6_-_44233499
|
1.705
|
NM_004556
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|
|
chr19_+_17326154
|
1.704
|
NM_018467
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae)
|
|
chr1_+_156698250
|
1.695
|
NM_001142560
NM_015997
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1
|
|
chr1_-_38471170
|
1.693
|
NM_001243878
NM_004468
|
FHL3
|
four and a half LIM domains 3
|
|
chr9_-_35749209
|
1.692
|
NM_020944
|
GBA2
|
glucosidase, beta (bile acid) 2
|
|
chr17_-_80009237
|
1.691
|
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr3_-_121740968
|
1.684
|
NM_001199799
NM_001199800
NM_175924
|
ILDR1
|
immunoglobulin-like domain containing receptor 1
|
|
chr6_-_31620468
|
1.678
|
NM_001199697
NM_001199698
NM_004639
NM_080702
NM_080703
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr12_-_6798619
|
1.669
|
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr21_+_46359896
|
1.666
|
NM_058190
|
FAM207A
|
family with sequence similarity 207, member A
|
|
chr19_+_17830302
|
1.664
|
NM_018174
|
MAP1S
|
microtubule-associated protein 1S
|
|
chrX_-_154255214
|
1.660
|
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr12_+_72057676
|
1.659
|
NM_031435
|
THAP2
|
THAP domain containing, apoptosis associated protein 2
|
|
chr19_-_7701852
|
1.656
|
|
|
|
|
chr17_-_80009649
|
1.656
|
NM_002917
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr16_+_67571980
|
1.632
|
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr12_-_6798419
|
1.626
|
|
ZNF384
|
zinc finger protein 384
|
|
chr22_+_38004676
|
1.622
|
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr19_-_46295785
|
1.608
|
NM_004943
|
DMWD
|
dystrophia myotonica, WD repeat containing
|
|
chr21_-_33104210
|
1.606
|
|
SCAF4
|
SR-related CTD-associated factor 4
|
|
chr8_+_144373534
|
1.605
|
NM_030895
|
ZNF696
|
zinc finger protein 696
|
|
chr7_+_18548899
|
1.598
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr6_+_31515391
|
1.590
|
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1
|
|
chr6_-_33290678
|
1.587
|
NM_001141969
NM_001141970
NM_001350
|
DAXX
|
death-domain associated protein
|
|
chr7_+_40174480
|
1.575
|
NM_001193311
NM_001193312
NM_001193313
NM_024728
|
C7orf10
|
chromosome 7 open reading frame 10
|
|
chr7_+_1570321
|
1.573
|
NM_002360
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr19_+_42387307
|
1.567
|
|
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1
|
|
chr22_+_38004865
|
1.562
|
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr6_-_33239585
|
1.557
|
NM_022553
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae)
|
|
chr19_+_39897457
|
1.556
|
NM_003407
|
ZFP36
|
zinc finger protein 36, C3H type, homolog (mouse)
|
|
chr19_-_46295647
|
1.549
|
|
DMWD
|
dystrophia myotonica, WD repeat containing
|
|
chr20_+_62289640
|
1.548
|
|
RTEL1
|
regulator of telomere elongation helicase 1
|
|
chr13_-_110438896
|
1.547
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr11_+_61129831
|
1.544
|
|
TMEM138
|
transmembrane protein 138
|
|
chr10_-_103454687
|
1.537
|
NM_022039
|
FBXW4
|
F-box and WD repeat domain containing 4
|
|
chr4_-_84376943
|
1.537
|
NM_133636
|
HELQ
|
helicase, POLQ-like
|
|
chr18_+_21033230
|
1.534
|
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr6_-_31620426
|
1.530
|
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr2_-_201753700
|
1.515
|
NM_032472
NM_130906
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3
|
|
chr19_+_17830283
|
1.513
|
|
MAP1S
|
microtubule-associated protein 1S
|
|
chr12_+_72057990
|
1.507
|
|
THAP2
|
THAP domain containing, apoptosis associated protein 2
|
|
chr17_+_18218608
|
1.502
|
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr6_-_31620384
|
1.501
|
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr18_+_21033285
|
1.493
|
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr2_-_69870976
|
1.488
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr1_-_155904174
|
1.487
|
|
KIAA0907
|
KIAA0907
|
|
chr9_-_35732389
|
1.475
|
NM_006289
|
TLN1
|
talin 1
|
|
chr16_+_3185037
|
1.473
|
NM_004220
|
ZNF213
|
zinc finger protein 213
|
|
chr3_+_150264514
|
1.473
|
NM_032025
|
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa
|
|
chr17_+_1945276
|
1.471
|
NM_080822
|
OVCA2
|
ovarian tumor suppressor candidate 2
|
|
chr19_+_34745452
|
1.462
|
NM_014686
|
KIAA0355
|
KIAA0355
|
|
chr22_-_39928651
|
1.460
|
|
RPS19BP1
|
ribosomal protein S19 binding protein 1
|
|
chr9_-_132404277
|
1.457
|
NM_001202403
NM_017873
NM_177999
|
ASB6
|
ankyrin repeat and SOCS box containing 6
|
|
chr17_-_62777621
|
1.456
|
|
LOC146880
|
Rho GTPase activating protein 27 pseudogene
|
|
chr1_-_155904201
|
1.454
|
|
KIAA0907
|
KIAA0907
|
|
chr19_-_1174201
|
1.453
|
|
|
|
|
chr3_-_40350948
|
1.447
|
|
FLJ33065
|
uncharacterized LOC440952
|
|
chr19_-_1174257
|
1.445
|
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr1_-_155904140
|
1.437
|
|
KIAA0907
|
KIAA0907
|
|
chr17_+_79633724
|
1.436
|
NM_199287
|
CCDC137
|
coiled-coil domain containing 137
|
|
chr15_-_74284518
|
1.424
|
NM_004809
|
STOML1
|
stomatin (EPB72)-like 1
|
|
chr12_+_52445185
|
1.422
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr17_+_18218605
|
1.422
|
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr3_-_128879873
|
1.418
|
NM_001204890
NM_001199469
NM_020701
|
ISY1-RAB43
ISY1
|
ISY1-RAB43 readthrough
ISY1 splicing factor homolog (S. cerevisiae)
|
|
chr4_+_103422553
|
1.415
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr11_-_45939361
|
1.410
|
|
PEX16
|
peroxisomal biogenesis factor 16
|
|
chr17_+_18218569
|
1.409
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr5_+_149380168
|
1.403
|
NM_014983
|
HMGXB3
|
HMG box domain containing 3
|
|
chr8_-_103666018
|
1.402
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr22_+_38004454
|
1.402
|
NM_001001560
NM_001001561
NM_001172687
NM_013365
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr17_-_79212884
|
1.398
|
NM_144679
|
C17orf56
|
chromosome 17 open reading frame 56
|
|
chr6_-_31634059
|
1.388
|
NM_001199237
|
GPANK1
|
G patch domain and ankyrin repeats 1
|
|
chr4_+_110736911
|
1.386
|
|
GAR1
|
GAR1 ribonucleoprotein homolog (yeast)
|
|
chr17_-_79876027
|
1.385
|
|
SIRT7
|
sirtuin 7
|
|
chr18_+_21033263
|
1.385
|
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr3_+_150264576
|
1.375
|
|
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa
|
|
chr4_-_75719686
|
1.372
|
NM_001729
|
BTC
|
betacellulin
|
|
chr20_+_62339321
|
1.369
|
NM_001195653
NM_001195654
NM_032527
NM_181485
|
ZGPAT
|
zinc finger, CCCH-type with G patch domain
|
|
chr18_+_3262110
|
1.368
|
NM_033546
|
MYL12B
|
myosin, light chain 12B, regulatory
|
|
chr1_-_38273683
|
1.365
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr16_-_67450149
|
1.362
|
NM_013304
|
ZDHHC1
|
zinc finger, DHHC-type containing 1
|
|
chr9_-_35748875
|
1.360
|
|
GBA2
|
glucosidase, beta (bile acid) 2
|
|
chr6_-_31633587
|
1.360
|
NM_001199238
NM_001199239
|
GPANK1
|
G patch domain and ankyrin repeats 1
|
|
chr9_-_35732175
|
1.357
|
|
TLN1
|
talin 1
|
|
chr10_-_99161013
|
1.354
|
NM_001145114
NM_015179
|
RRP12
|
ribosomal RNA processing 12 homolog (S. cerevisiae)
|
|
chr9_-_35732115
|
1.354
|
|
TLN1
|
talin 1
|
|
chr1_-_38273852
|
1.352
|
NM_024640
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr9_+_35732629
|
1.348
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr16_-_21831730
|
1.347
|
|
RRN3P1
|
RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene 1
|
|
chr11_-_64885124
|
1.342
|
NM_014205
|
ZNHIT2
|
zinc finger, HIT-type containing 2
|
|
chr12_-_6798528
|
1.339
|
NM_133476
|
ZNF384
|
zinc finger protein 384
|
|
chr6_-_11382501
|
1.337
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chrX_+_47092362
|
1.337
|
|
USP11
|
ubiquitin specific peptidase 11
|
|
chrX_-_48937560
|
1.336
|
NM_001029896
|
WDR45
|
WD repeat domain 45
|
|
chr15_-_64648214
|
1.334
|
|
CSNK1G1
|
casein kinase 1, gamma 1
|
|
chr9_-_123691442
|
1.333
|
NM_001190945
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr6_-_31620142
|
1.329
|
NM_001098534
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr7_+_18548963
|
1.322
|
|
HDAC9
|
histone deacetylase 9
|
|
chr12_+_122356446
|
1.321
|
NM_001178003
NM_144668
|
WDR66
|
WD repeat domain 66
|
|
chr5_+_172483285
|
1.320
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr1_-_155904186
|
1.318
|
NM_014949
|
KIAA0907
|
KIAA0907
|
|
chr16_-_54319966
|
1.317
|
|
IRX3
|
iroquois homeobox 3
|
|
chr12_-_57914287
|
1.316
|
NM_001195053
NM_001195054
NM_001195055
NM_001195056
NM_001195057
NM_004083
|
DDIT3
|
DNA-damage-inducible transcript 3
|
|
chr1_-_38273846
|
1.310
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr9_-_35732089
|
1.306
|
|
TLN1
|
talin 1
|
|
chr9_-_35732226
|
1.305
|
|
TLN1
|
talin 1
|
|
chr16_+_284798
|
1.303
|
NM_032039
|
ITFG3
|
integrin alpha FG-GAP repeat containing 3
|
|
chr16_+_10837673
|
1.301
|
NM_002484
|
NUBP1
|
nucleotide binding protein 1
|
|
chr17_-_79269082
|
1.300
|
NM_001037984
NM_138570
|
SLC38A10
|
solute carrier family 38, member 10
|
|
chr17_+_62502860
|
1.294
|
|
CEP95
|
centrosomal protein 95kDa
|
|
chr5_+_149380310
|
1.292
|
|
HMGXB3
|
HMG box domain containing 3
|
|
chr17_-_79212775
|
1.291
|
|
C17orf56
|
chromosome 17 open reading frame 56
|
|
chr5_+_173315313
|
1.286
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr16_-_66968265
|
1.283
|
NM_016062
|
FAM96B
|
family with sequence similarity 96, member B
|
|
chr22_+_20861964
|
1.281
|
|
MED15
|
mediator complex subunit 15
|
|
chrX_+_47092264
|
1.278
|
NM_004651
|
USP11
|
ubiquitin specific peptidase 11
|
|
chr8_+_22102618
|
1.278
|
NM_001722
|
POLR3D
|
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa
|
|
chr16_+_2097985
|
1.276
|
NM_000548
NM_001077183
NM_001114382
|
TSC2
|
tuberous sclerosis 2
|
|
chr10_-_99160987
|
1.271
|
|
RRP12
|
ribosomal RNA processing 12 homolog (S. cerevisiae)
|
|
chr14_-_75530716
|
1.268
|
NM_001107
NM_203488
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type
|
|
chr9_+_130830447
|
1.267
|
NM_001006641
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr5_+_180650709
|
1.266
|
|
TRIM41
|
tripartite motif containing 41
|
|
chr13_+_73356179
|
1.266
|
NM_006346
|
PIBF1
|
progesterone immunomodulatory binding factor 1
|
|
chr2_+_220408352
|
1.262
|
|
TMEM198
|
transmembrane protein 198
|
|
chr7_+_157129659
|
1.262
|
NM_005494
NM_058246
|
DNAJB6
TMEM135
|
DnaJ (Hsp40) homolog, subfamily B, member 6
transmembrane protein 135
|
|
chr21_-_33765149
|
1.261
|
NM_014825
|
URB1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae)
|
|
chr4_+_75480628
|
1.260
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr17_+_43299277
|
1.257
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr19_+_10216898
|
1.255
|
NM_020230
NM_001040664
NM_001198690
|
PPAN
PPAN-P2RY11
|
peter pan homolog (Drosophila)
PPAN-P2RY11 readthrough
|
|
chrX_+_49091979
|
1.251
|
|
CCDC22
|
coiled-coil domain containing 22
|
|
chr12_-_112037360
|
1.238
|
NM_002973
|
ATXN2
|
ataxin 2
|
|
chr9_-_3525973
|
1.237
|
NM_002919
NM_134428
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr1_-_228594466
|
1.236
|
NM_145214
|
TRIM11
|
tripartite motif containing 11
|
|
chr4_+_109088677
|
1.233
|
|
LOC641518
|
uncharacterized LOC641518
|
|
chr7_+_1609737
|
1.232
|
|
KIAA1908
|
uncharacterized LOC114796
|
|
chr5_-_6378595
|
1.227
|
NM_032286
|
MED10
|
mediator complex subunit 10
|
|
chr7_+_149412027
|
1.227
|
NM_032534
|
KRBA1
|
KRAB-A domain containing 1
|
|
chr2_-_220041549
|
1.225
|
NM_015680
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1
|
|
chr4_+_103422479
|
1.216
|
NM_001165412
NM_003998
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr15_-_41836458
|
1.215
|
NM_015540
|
RPAP1
|
RNA polymerase II associated protein 1
|
|
chr18_-_33077896
|
1.214
|
NM_001098817
NM_194281
|
INO80C
|
INO80 complex subunit C
|
|
chr13_+_46039024
|
1.212
|
NM_031431
|
COG3
|
component of oligomeric golgi complex 3
|
|
chr17_+_45973515
|
1.210
|
NM_003110
|
SP2
|
Sp2 transcription factor
|
|
chr11_+_47430045
|
1.208
|
NM_001128225
NM_152264
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr6_-_33239439
|
1.207
|
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae)
|